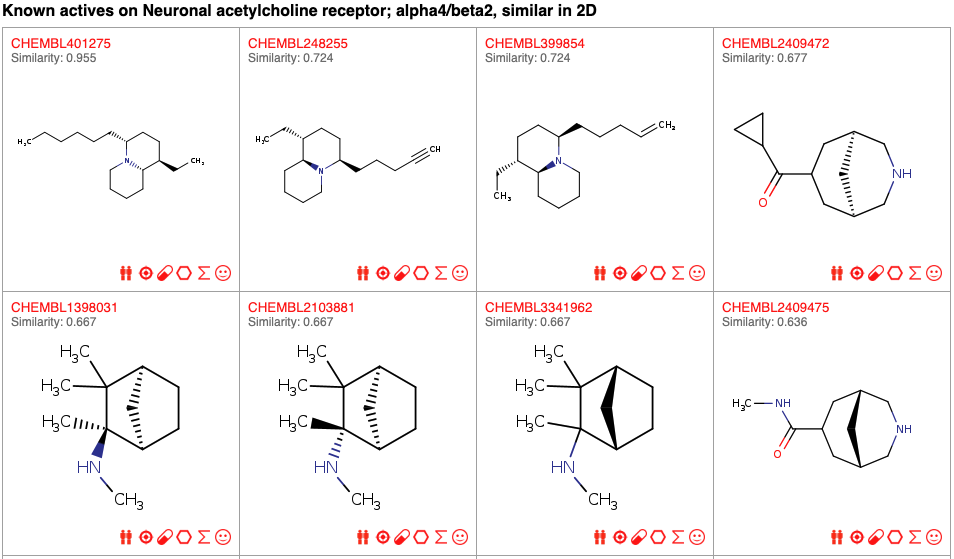
Known Actives Page
This page displays the compounds, which have driven the prediction for a given protein as target of the query molecule. These compouds are experimentally defined as actives on the target under investigation (here, the subunits alpha4 and beta2 of the neuronal acetylcholine receptor) and highly similar to your query molecule based on 2D (in this example) or on 3D simlarity measures. Each active is identified by its Chembl ID (clickable link to ChEMBL) and similarity values are indicated below.When predictions are made by homology (i.e., compounds most similar to the query molecule are known actives on a homolog of the predicted target), the actual protein target is also shown.
Each compound is accompagnied with interoperability icons to enable straightforward submission to different in house web tools (e.g. SwissSimilarity or SwissADME).
Clickable snapshot